HARWOOD GENOMICS
Comparative genomics of environmental stress responses in North American hardwoods
Funded by NSF (National Science foundation)
February 1, 2011 To January 31, 2015
Website: http://www.hardwoodgenomics.org/
Executive summary
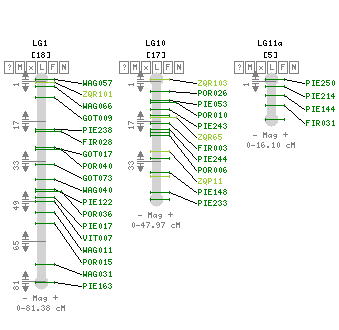
This project, entitled "Comparative genomics of environmental stress responses in North American hardwoods", is creating genomic resources for an array of the eight most economically and phylogenetically important hardwood species in North America. This includes EST databases for 6 species, BAC libraries for 6 species, framework genetic maps for 3 species, and high density QTL maps for 2 species. Funded by the National Science Foundation’s Plant Genome Research Program, this multi-institutional award is being lead by Dr. John Carlson from Pennsylvania State University in collaboration with scientists at six additional Universities to meet research and outreach goals.
Data produced by the project is being made available to others as quickly as it can be checked and validated. Announcements of new data and links to download/explore this data will be available on this site.
Species and resources being studied:
-
Sweetgum (EST, gSSRs, Framework Map, Reference Populations)
-
Honeylocust (EST, gSSRs, Framework Map, Reference Populations)
-
Black Walnut (EST, gSSRs, BAC, QTL Map, Refereunce Populations)
-
Northern Red Oak (EST, BAC, QTL Map, Reference Populations, Gene Space Physical Map)
-
Sugar Maple (EST, gSSRs)
-
Green Ash (EST, gSSRs)
-
Tulip Poplar (Framework Map, Reference Populations)
This site will also integrate existing resources from other hardwood tree species. These species include:
-
American Beech (EST)
-
Chinese Chestnut (EST, BAC, QTL Map, Whole Genome Sequence)
-
American Chestnut (EST, BAC, Framework Map)
-
Pedunculate Oak (EST, BAC, QTL Map)
-
White Oak (EST)