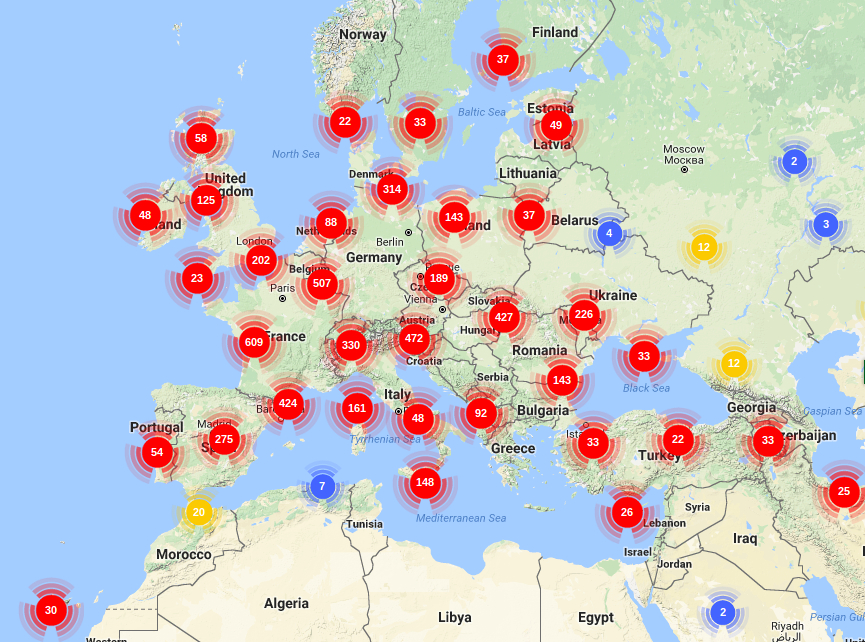
QUERCUS EST DATABASE
This page provides access to either dedicated databases, publications or flat files for three unigene sets constructed for the genus Quercus :
BMC Genomics 11: 650), the oak unigene set contains 1,704,117 ESTs collapsed into 69,154 tentative contigs and 153,517 singletons, providing 222,671 non-redundant sequences (including alternative transcripts). Clustering and assembly were performed using the TGICL pipeline.
In its first version Oakcontig v1 (Ueno et al. 2010:
OakContigDF159.1, a reference library for studying differential gene expression in Quercus robur during controlled biotic interactions: use for quantitative transcriptomic profiling of oak roots in ectomycorrhizal symbiosis (Tarkka et al. 2013: New Phytologist 199: 529-540)
OCV3: de novo assembly of the oak transcriptome
These data are associated with the following publication:
The oak gene expression atlas: Insights into Fagaceae genome evolution and the discovery of genes regulated during bud dormancy release
Lesur I, Le Provost G, Bento P, Da Silva C, , Leplé JC, Murat F, Ueno S, Bartholomé J, Lalanne C, Ehrenmann F, Noirot C, Burban C, Loustau ML, Léger V, Amselem J, Belser C, Quesneville H, Stierschneider M, Fluch S, Feldhahn L, Tarkka M, Herrmann S, Buscot F, Klopp C, Kremer A, Salse J, Aury JM, Plomion C (2015) The oak gene expression atlas: insights into Fagaceae genome evolution and the discovery of genes regulated during bud dormancy release. BMC Genomics 16:112
Oak contig V3 (OCV3-91k and OCV3-101k, FASTA format, zip archive) :
OCV3_assembly.fasta