OAK GENOME SEQUENCE
Website : http://www.oakgenome.fr
An international consortium has recently sequenced the genome of the pedunculate oak. The work (Plomion et al. 2018) reveals two main genomic features related to the longevity of this emblematic species. The first is the existence in the genome of a particularly rich and diverse set of genes involved in resistance against pests (fungi, oomycetes, insects, bacteria and viruses). This extended defense arsenal affords trees protection against their dominant predators and is likely a key to their longevity. The second feature revealed by the work is the existence of somatic mutations that can be transmitted to the next generation - a result that raises questions about the importance of somatic mutations (non germ line) in generating genetic diversity in long-lived species.
A reference genome for one of the 400 oak species
The pedunculate oak (Quercus robur) genome was sequenced using high throughput sequencing technologies (Plomion et al. 2018). More specifically, a highly contiguous haploid genome (PM1N, table 1) was generated based on Illumina synthetic long-reads and 454 (Roche) sequences. The assembly contains 1,409 scaffolds totaling 814 Mb (N50, 1.34 Mb). Overall 871 scaffolds, representing 96% of the physical size of the genome, were anchored to the 12 chromosomes. Three other draft genomes, while of lower quality, have also been published (Sork et al. 2016; Schmid-Siegert et al. 2017 and Ramos et al. 2018) (Table 2).
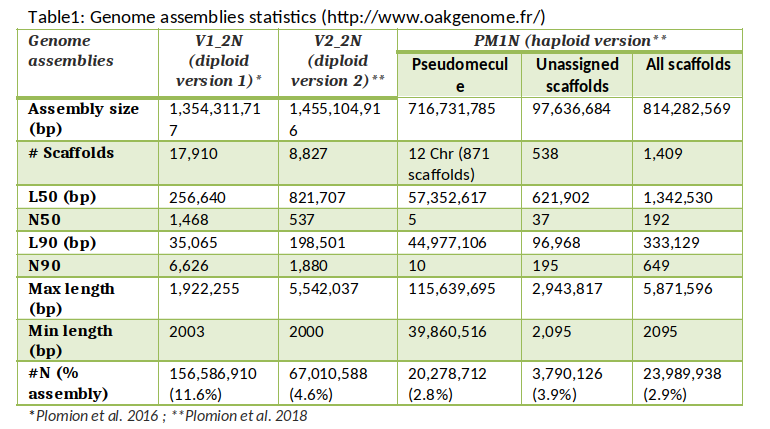
The assembly contains 94,394 scaffolds, totaling 1.17 Gb with 18,512 scaffolds of length 2 kb or longer, with a total length of 1.15 Gb, and a N50 scaffold size of 278,077 kb. The k-mer histograms indicate an diploid genome size of ∼720–730 Mb, which is smaller than the total length due to high heterozygosity, estimated at 1.25%. A comparison with a recently published European oak (Q. robur) nuclear sequence indicates 93% similarity. The Q. lobata chloroplast genome has 99% identity with another North American oak, Q. rubra. Preliminary annotation yielded an estimate of 61,773 predicted protein-coding genes, of which 71% had similarity to known protein domains. We searched 956 Benchmarking Universal Single-Copy Orthologs, and found 863 complete orthologs, of which 450 were present in > 1 copy.
A genome with an arsenal of defense genes against biotic threats, a likely key to longevity.
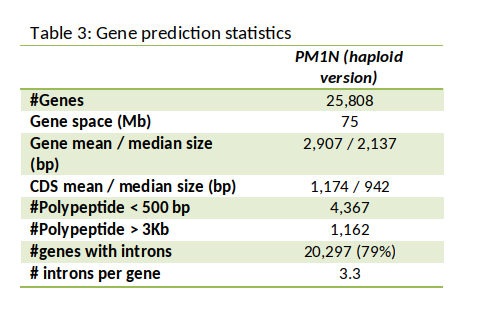
The annotation of the Pedunculate oak genome showed that it is enriched with transposable elements at 51% of total DNA and contains 25,808 genes (Table 2). Among these genes, 36% are organized in clusters of continuous genes (so called tandem duplicates); this number is much higher than found in other plants at ~15 %. It was also shown that the number of resistance genes increased mostly via tandem duplication. The comparison between the genomes of herbaceous species (Arabipdopsis, soybean, potato, watermelon…) and perennial woody species (oak, poplar, eucalyptus, peach tree…) indicated that this expansion of resistance genes is not specific to oaks, but shared among tree species. As trees remain living across years, they are continually barraged by pests; these pests likely evolve faster than their tree hosts due to their much shorter generation times. In these conditions the diversity and richness of immunity genes could help trees to cope with a broad range of short-lived microbial enemies and help them to persist and live longer.
Are trees genomic mosaics ?
Multi-cellular organisms accumulate somatic mutations as they grow. The long life of trees, often reaching hundreds of years for some species, and their continuous growth across their lives makes them perfect models to investigate this phenomenon. In their paper, Plomion et al. (2018) investigated the frequency of somatic mutations by comparing the genomes of samples harvested at the extremities of branches of different ages on a centennial oak tree. This strategy allowed to identify rare somatic mutations (as also found by Schmid-Siegert et al. 2017), but also to show that they can be transmitted to the next generation. Whether some of these mutations can confer individuals with a selective edge remains to be studied.