OAK-ADAPT
Séquençage haut-débit de génomes et de transcriptomes de chêne sessiles afin de mieux comprendre les phénomènes d’adaptation de cette espèce à son environnement.
Funded by ANR investissement d'avenir France Génomique
01 February 2014 to 31 December 2015
Website: https://www.france-genomique.org/spip/spip.php?article114&lang=fr
Partner labs: URGI (Versailles), CNRGV (Toulouse), MIA-T (Toulouse), GDEC (Clermont-Ferrand), UMR BIOGECO (Bordeaux-France).
Coordinator: Christophe Plomion (UMR Biogeco) christophe.plomion@inra.fr
Executive summary
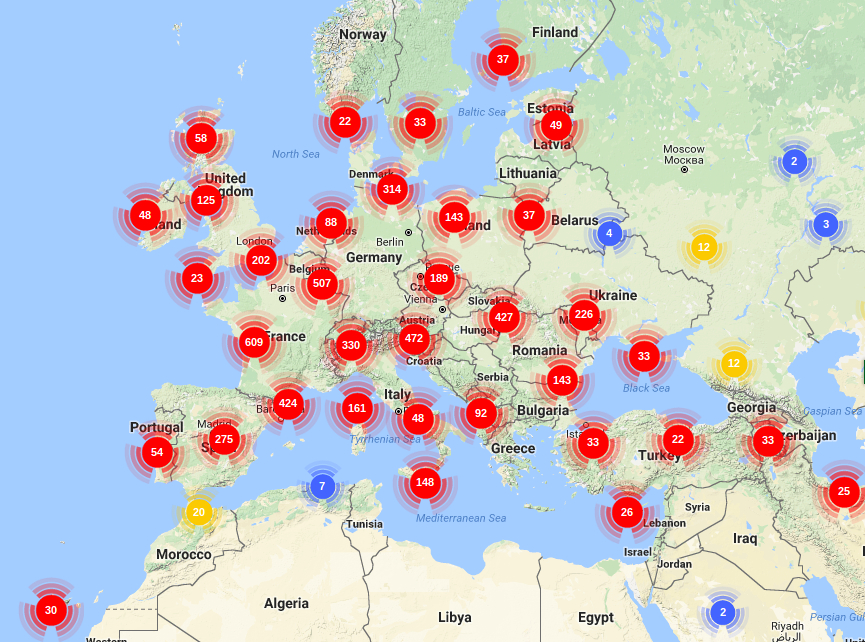
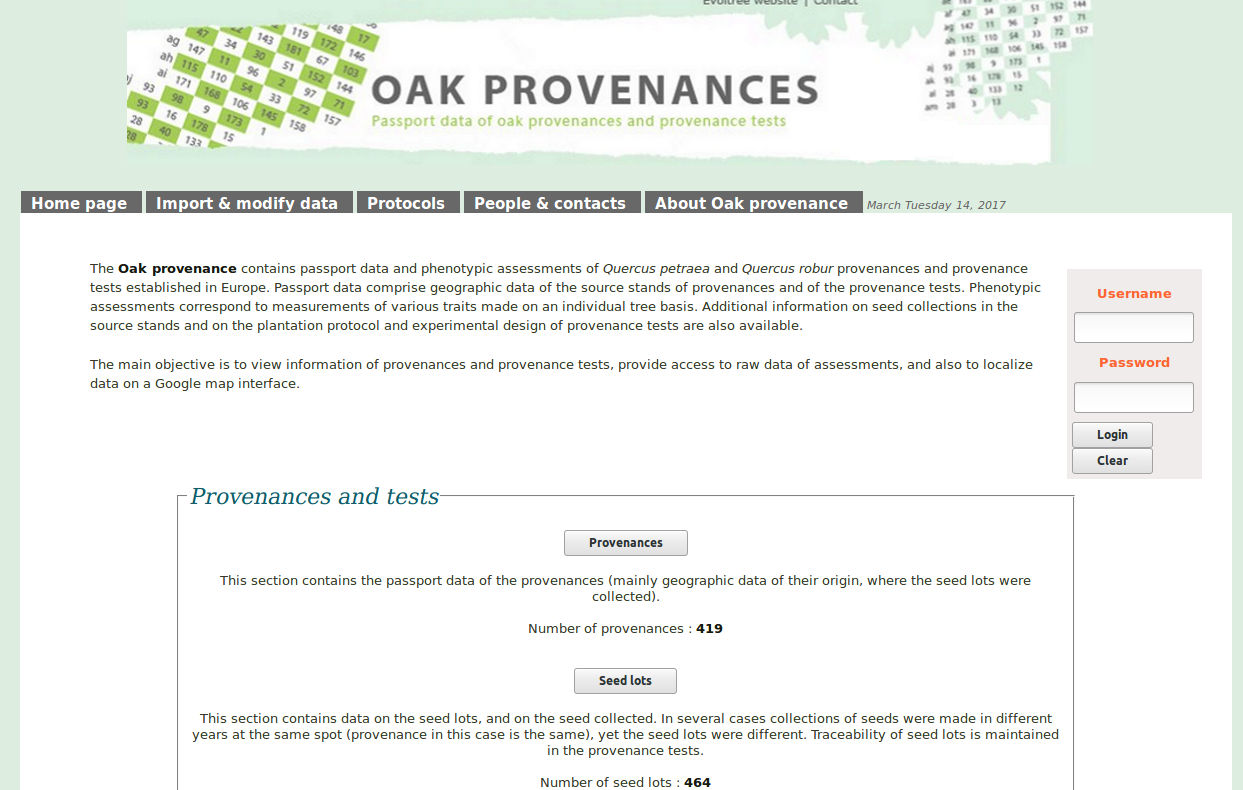
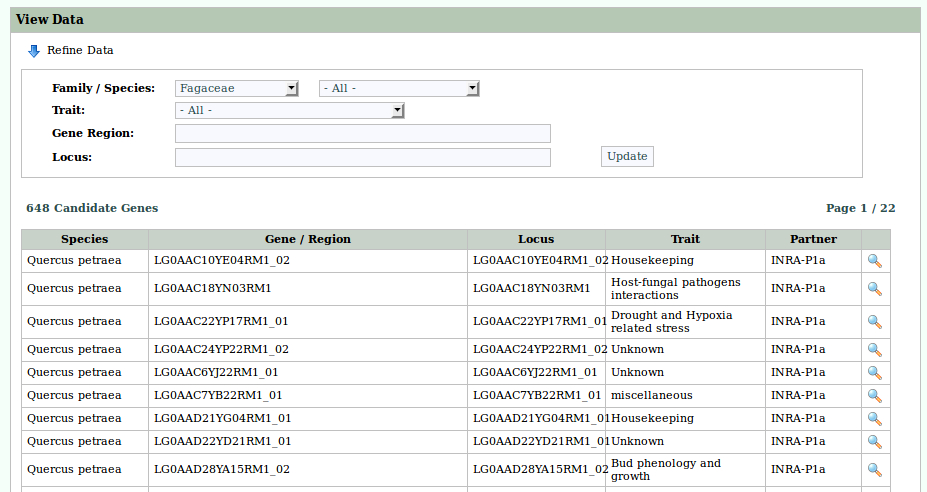
The main objective of this project is to explore the role of gene expression and structural variation in contributing to an adaptive traits (bud phenology) in a keystone forest tree species: oak.
- We will generate sequences (RNAseq and pool-seq data) at the population level to study local adaptation, a process whereby a population becomes better suited to its habitat. The selected populations are located along two elevation clines in the Pyrenees, showing strong phenotypic differences in terms of average bud burst date.
- We will also generate sequences within a structured populations (endo- and eco-dormant buds sampled on 150 full-sibs) to decipher the genetic architecture of gene expression and how it relates to bud phenology QTLs.