OAK GENOME SEQUENCING
GENOAK : Sequencing of the oak genome and identification of genes that matter for forest tree adaptation
Funding agencies: INRA (1.2M€), CEA (0.4M€), ANR (0.67M€) - Starting date: 1st nov. 2011
Website: OAK GENOME SEQUENCING 
 Genoak users access (Login)
Genoak users access (Login)
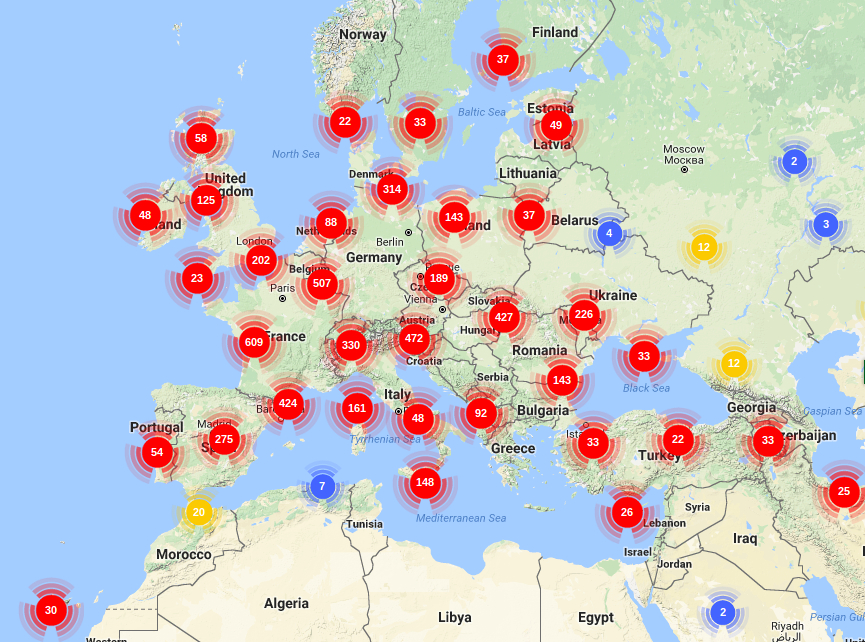
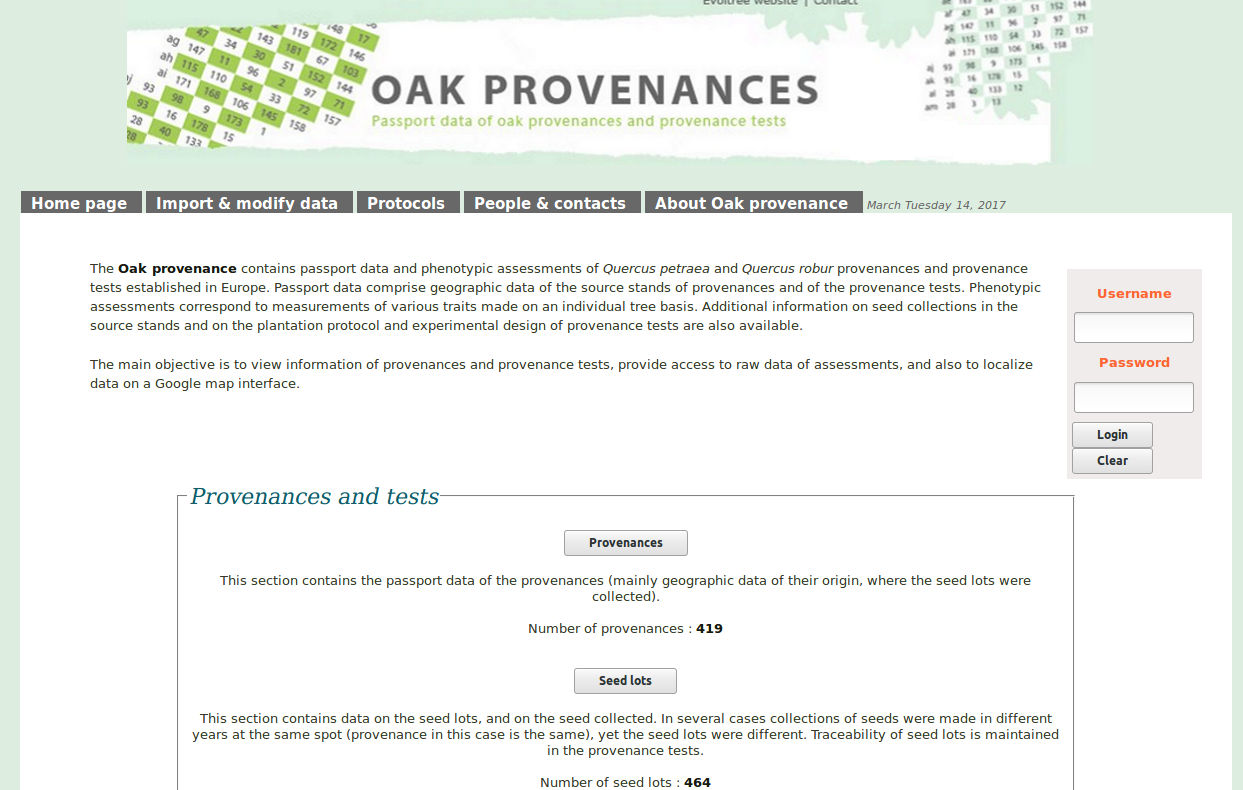
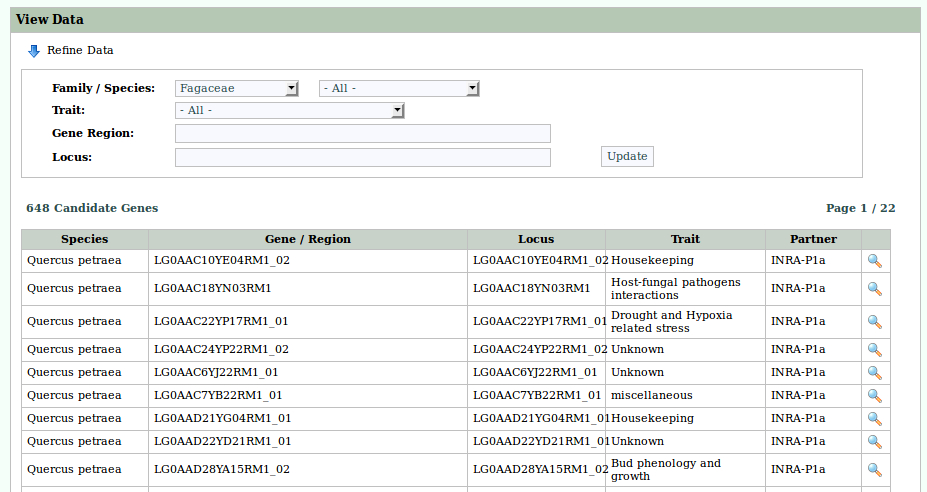
Trees are an important component of the global ecosystem. They dominate many regions of the world as natural, extensively or intensively managed plantations. The Fagaceae family comprises about 1,000 woody species distributed throughout the northern hemisphere. About half belong to the Quercus (oak) genus. These oaks have traditionally high cultural and societal values. As other forest tree species, they are also providing important environmental (carbon sequestration, water cycle, reservoir of biodiversity, soil protection...) and economic (carpentry, furniture, cabinet making, veneer, cask industry, fuelwood, hunting and fungus gathering) services. Finally these long-lived organisms are considered as relevant models to study both short and long term adaptation mechanisms to abiotic and biotic constraints associated with global change, as they grow under a wide range of environments (soil and climate). Due to their biological dominance, their commercial and ecological importance worldwide, an international network was launched recently to develop a coordinated strategy to sequence several Fagaceae genomes. These genomic resources will allow to discover which genes are involved in the adaptation of these organisms to their environments and to study their adaptive potential under current and future climate changes. In Europe, genomic resources have been developed for sessile and pedunculate oaks in the frame of the EVOLTREE network of excellence. In US, American and Chinese chestnuts (Castanea) were the main targets of a NSF funded project. Their genome is now being sequenced within the Forest Health Initiative. The goal is to provide a high-quality reference sequence for the Castanea mollissima (Chinese Chestnut) genome, with which to identify genes for resistance to Cryphonectria parasitica. In China, the efforts to sequence the Lithocarpus and Castanopsis genomes are also on the right track. The present project aims at supporting the participation of French Institutions involved in this international effort. It is a collaboration between four INRA research units and the GENOSCOPE: the French National Sequencing Center.
The objective of the GENOAK project are two-fold:
- establish a high quality reference genome sequence for Quercus robur (740Mb/C), anchored to an ultra high density SNP-based linkage map on which a series of trait-QTLs have been localized. This resource will provide genetic links between traits and sequence variations at a high resolution,
- identify genes involved in adaptation through the analysis of adaptive divergence (incipient ecological speciation) at the transcriptome and genome-wide levels by respectively i) identifying differentially expressed genes between co-occuring species (sessile and pedunculate oaks) adapted to different edapho-climatic environments, and ii) analyzing the size, distribution and nature of genomic islands of divergence maintained in the face of gene flow of the same sympatric species.
Tasks
- obtain a reference genome for a single Pedunculate oak genotype
- assign genome scaffolds on the genome using a highly saturated SNP-based genetic linkage map
- annotate structurally and functionally the genome
- integrate the whole genome with automatic prediction into a Genome Browser
- study the micro-synteny and colinearity between oak and chestnut, as well as with other eudicots, for which a genome sequence is available
- reveal genetic variations that are responsible for important adaptive traits in oaks by resequencing the genome of several individuals of two sister species.

other associated partners: P Faivre-Rampant (INRA Evry), C Klopp and H Berges (INRA Toulouse)
Recent publications on oak genomics
- Plomion C, Aury JM, Amselem J, Alaeitabar T, Barbe V, Belser C, Bergès H, Bodénès C, Boudet N, Boury C, Canaguier A, Couloux A, Da Silva C, Duplessis S, Ehrenmann F, Estrada-Mairey B, Fouteau S, Francillonne N, Gaspin C, Guichard C, Klopp C, Labadie K, Lalanne C, Le Clainche I, Leplé JC, Le Provost G, Leroy T, Lesur I, Martin F, Mercier J, Michotey C, Murat F, Salin F, Steinbach D, Faivre-Rampant P, Wincker P, Salse S, Quesneville H, Kremer A (2015) Decoding the oak genome: public release of sequence data, assembly, annotation and publication strategies. Mol Ecol Res. DOI: 10.1111/1755-0998.12425
- Lepoittevin C, Bodénès C, Chancerel E, Villate L, Lang T, Lesur I, Boury C, Ehrenmann F, Zelenica D, Boland A, Besse C, Garnier-Gérè P, Plomion C, Kremer A (2015) Single-nucleotide polymorphism discovery and validation in high density SNP array for genetic analysis in European white oaks. Mol Ecol Res doi: 10.1111/1755-0998.12407
- Goicoechea PG, Herran A, Durand J, Bodénès C, Plomion C, Kremer A (2015) SA linkage disequilibrium perspective on the genetic mosaic of speciation in two hybridizing Mediterranean white oaks. Heredity 114: 373-86
- Lesur I, Le Provost G, Bento P, Da Silva C, , Leplé JC, Murat F, Ueno S, Bartholomé J, Lalanne C, Ehrenmann F, Noirot C, Burban C, Loustau ML, Léger V, Amselem J, Belser C, Quesneville H, Stierschneider M, Fluch S, Feldhahn L, Tarkka M, Herrmann S, Buscot F, Klopp C, Kremer A, Salse J, Aury JM, Plomion C (2015) The oak gene expression atlas: insights into Fagaceae genome evolution and the discovery of genes regulated during bud dormancy release. BMC Genomics 16:112
- Lesur I, Bechade A, Lalanne C, Klopp C, Noirot C, Leplé JC, Kremer A, Plomion C, Le Provost G (2015) A unigene set for European beech (Fagus sylvatica L.) and its use to decipher the molecular mechanisms involved in dormancy regulation. Mol Ecol Res 15:1192-1204
- Gailing O, Bodénès C, Finkeldey R, Kremer A, Plomion C (2013) Genetic mapping of EST-derived Simple Sequence Repeats (EST-SSRs) to identify QTL for leaf morphological characters in a Quercus robur full-sib family. Trees Genetics & Genomes 9: 1361-1367
- Plomion C, Fievet V (2013) Oak genomics takes off and enters the ecological genomics era. New Phytol 199: 308-31
- Petit RJ, Carlson J, Curtu L, Loustau ML, Plomion C, Rodriguez AG, Sork V, Ducousso A (2013). Fagaceae trees as models to integrate ecology, evolution and genomics. New Phytol 197: 369-371
- Ueno S, Klopp C, Noirot C, Léger V, Prince E, Kremer A, Plomion C, Le Provost G (2013) Transcriptional profiling of bud dormancy induction and release in oak by next-generation sequencing. BMC Genomics 14:236
- Bodénès C, Chancerel E, Murat F, Bagnoli F, Gailing O, Durand J, Koelewijn HP, Goicoechea P, Villani F, Roussel G, Salse S, Vendramin GG, Soularue JP, Boury C, Alberto F, Kremer A, Plomion C (2012) Comparative mapping in the Fagaceae and beyond using EST-SSRs. BMC Plant Biol 12:153
- Kremer A, Abbott AG, Carlson JE, Manos PS, Plomion C, Sisco P, Staton ME, Ueno S, Vendramin GG (2012) Genomics of Fagaceae. Trees Genetics & Genomes. Trees Genetics & Genomes (2012) 8:583-610
- Le Provost G, Sulmon C, Frigério JM, Bodénès C, Kremer A, Plomion C (2012) The role of water-logging responsive genes in shapping inter-specific differentiation between two sympatric oak species. Tree Physiol 32: 19-134
- Lesur I, Durand J, Sebastiani F, Gyllenstrand N, Bodénès C, Lascoux M, Kremer K, Vendramin GG, Plomion C (2011) A sample view of the pedunculate oak (Quercus robur) genome from the sequencing of hypomethylated and random genomic libraries. Trees Genetics & Genomes 7: 1277-1285
- Vornam B, Gailing O, Dérory J, Plomion C, Kremer A, Finkeldey R (2011) Characterization and allelic variation of a dehydrin gene in Quercus petraea (Matt.) Liebl. Plant Biol 13: 881-887
- Faivre-Rampant P, Lesur I, Boussardon C, Bitton F, Bodénès C, Le Provost G, Bergès H, Fluch S, Kremer A, Plomion C (2011). Analysis of BAC end sequences in oak, providing insights into the composition of the genome of this keystone species. BMC Genomics 12: 292
- Durand J, Bodénès C, Chancerel E, Frigerio JM, Vendramin G, Sebastiani F, Buonamici A, Gailing O, Koelewijn HP, Villani F, Mattioni C, Cherubini M, Goicoechea PG, Herrán A, Ikaran Z, Cabané C, Ueno S, Alberto F, Dumoulin P-Y, Guichoux E, de Daruvar A, Kremer A, Plomion C (2010) A fast and cost-effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genomics 11: 570
- Ueno S, Le Provost G, Léger V, Klopp C, Noirot C, Frigerio J-M, Salin F, Salse J, Abrouk M, Murat F, Brendel O, Derory J, Abadie P, Léger P, Cabane C, Barré A, de Daruvar A, Couloux A, Wincker P, Reviron M-P, Kremer A, Plomion C (2010) Bioinformatic analysis of ESTs collected by Sanger and pyrosequencing methods for a keystone forest tree species: oak. BMC Genomics 11: 650