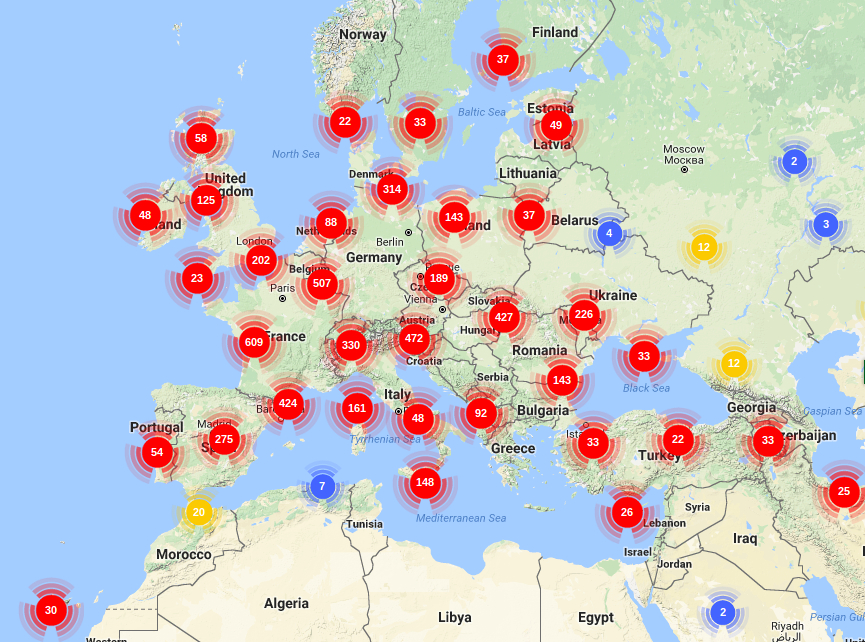
OCV1 UNIGENE SETS
Direct Link: http://genotoul-contigbrowser.toulouse.inra.fr
The EST database contains the most comprehensive EST collection of oaks (Quercus petraea, Q. robur and Q. suber). Sequences were generated using the Sanger and 454 Roche methods from 40 cDNA libraries.
In its first version Oakcontig v1 (Ueno et al. 2010: BMC Genomics 11: 650), the oak unigene set contains 1,704,117 ESTs collapsed into 69,154 tentative contigs and 153,517 singletons, providing 222,671 non-redundant sequences (including alternative transcripts). Clustering and assembly were performed using the TGICL pipeline.
Gene ontology annotation was assigned to 29,303 unigene elements. Blast search against the SWISS-PROT database revealed putative homologs for 32,810 (14.7 %) unigene elements, but more extensive search with Pfam, Refseq_protein, Refseq_RNA and eight gene indices revealed homology for 67.4 % of them. The EST catalogue was examined for putative homologs of candidate genes involved in bud phenology, cuticle formation, phenylpropanoids biosynthesis and cell wall formation. Our results suggest a good coverage of genes involved in these traits.
Simple sequence repeats (SSRs) and single nucleotide polymorphisms (SNPs) were searched, resulting in 52,834 SSRs and 36,411 SNPs.
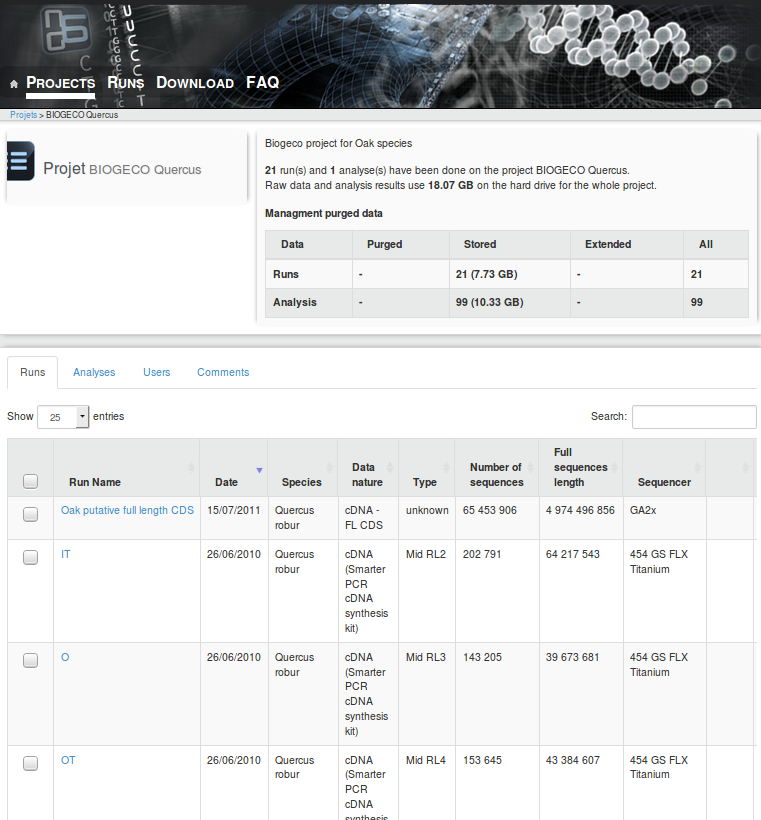
Access to the raw data : http://ng6.toulouse.inra.fr/ (Login: biogeco Password: 33bio31geno?)
*Raw data have been released in public databases :
-Sanger ESTs: http://www.ncbi.nlm.nih.gov/sites/entrez
Dec. 2010
Quercus mongolica subsp. crispula (3385)
-454-reads: http://www.ncbi.nlm.nih.gov/sra
(Dec. 2010, 24 libraries)
*cDNA clones are available at http://www.evoltree.eu/index.php/repository-centre